What to Consider When Planning a Gene Knockout Experiment with CRISPR/Cas9
This tutorial is part 1 of 4 in the series "How to create a gene knockout using CRISPR." Use Benchling's free molecular biology tools to plan your own CRISPR experiment and design your own gRNAs here.
Introduction
When generating a gene knockout, CRISPR is the easiest, cheapest and fastest gene editing technique. Here we provide a comprehensive, step-by-step tutorial to help you design your first CRISPR gene knockout experiment. Our series will cover how you can:
Generate a Knockout Using CRISPR: The Mechanism
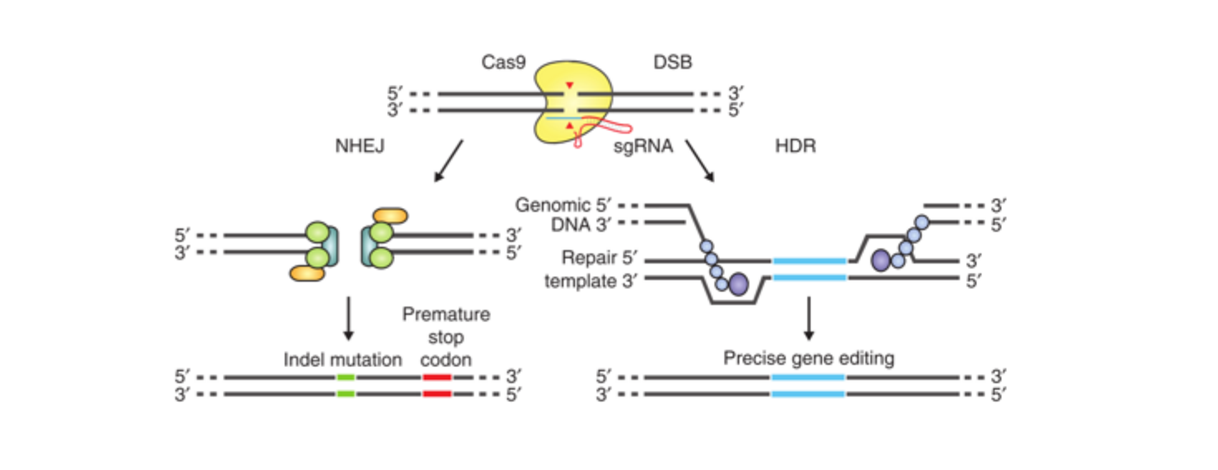
Before we get into the experimental details, let’s review how CRISPR works: CRISPR consists of a guide RNA (gRNA) and a DNA endonuclease, such as Cas9. The gRNA determines where gene knockouts will occur. These knockouts are also called insertions or deletions (indels). Once the gRNA and Cas9 are expressed in cells, the gRNA will direct Cas9 to bind to the target sequence and introduce a double-strand break. The cell will repair the break with either non-homologous end joining (NHEJ) or homolog directed repair (HDR).
NHEJ is the most active repair mechanism but is often inaccurate, and can lead to mutations, or indels, in the genetic code. If the indel occurs within the open reading frame, this insertion or deletion of genetic material, when transcribed, will render the gene non-functional. This is otherwise known as a "gene knockout." You can then use mismatch cleavage assays to identify which cells contain these indels at your gene of interest. By using the cell’s imperfect repair mechanism, CRISPR allows you to simply construct cell lines with knockouts at your genomic region of choice.
Key Considerations for Your CRISPR Experiment
Before you start, you need to determine:
Which cell line to work with:Is your cell line hard to transfect (e.g. Jurkat, primary cell lines) or post-mitotic (e.g. neurons, hepatocyte)? In this case, you might have a higher delivery efficiency using viral vectors, rather than chemical transfection. However, viral production can be difficult and time-consuming. If you are new to the protocol, it could take a month to optimize the procedure.
Which endonuclease to use:Streptococcus pyogenesCas9 (SpCas9) is the most common Cas for genome engineering. But you might still want to consider other variants if:
You want to use AAV to deliver CRISPR:The maximal cargo size for AAV is around 4.5 kb and just SpCas9 itself is 4.2 kb. If you need to add control elements or selection markers to your CRISPR, then a smaller Cas9, such as Staphylococcus aureusCas9 (SaCas9) or Streptococcus thermophilus Cas9 (St1Cas9) might work better for you.
Your target genomic region does not have the SpCas9 protospacer adjacent motif (PAM) sequence, 5’-NGG-3’: The target sequence has to be immediately upstream of the PAM sequence to ensure successful Cas9 binding. There are other Cas9 variants with different PAM sequences (Table 1). If your target sequence is AT-rich, Cpf1, another RNA-guided DNA endonuclease, could be a good choice for you.
You need to reduce further unwanted off-target mutations:Researchers have modified SpCas9 to increase its specificity; examples include SpCas9 D1135E, eSpCas9, and SpCas9-HF1. These variants are new and have not been as widely used as wild type SpCas9. But the research backing them is sound. They might be worth a try.
What’s next?
OK. Now you have decided the cell line and Cas9 for your experiments. In the next blog post, we will show you how to design your gRNAs.
Want to use Benchling's CRISPR tools at your company?
References
Jinek, Martin et al."A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity." Science 337.6096 (2012): 816-821. ^
Cong, Le et al."Multiplex genome engineering using CRISPR/Cas systems." Science 339.6121 (2013): 819-823. ^
Mali, Prashant et al."RNA-guided human genome engineering via Cas9." Science 339.6121 (2013): 823-826. ^
Ran, F Ann et al."In vivo genome editing using Staphylococcus aureus Cas9." Nature 520.7546 (2015): 186-191. ^
Esvelt, Kevin M et al. "Orthogonal Cas9 proteins for RNA-guided gene regulation and editing." Nature methods 10.11 (2013): 1116-1121. ^
Zetsche, Bernd et al."Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system." Cell 163.3 (2015): 759-771. ^
Kleinstiver, Benjamin P et al."Engineered CRISPR-Cas9 nucleases with altered PAM specificities." Nature (2015). ^
Slaymaker, Ian M et al."Rationally engineered Cas9 nucleases with improved specificity." Science 351.6268 (2016): 84-88. ^
Kleinstiver, Benjamin P et al."High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects." Nature (2016). ^
Ran, F Ann et al."In vivo genome editing using Staphylococcus aureus Cas9." Nature 520.7546 (2015): 186-191. ^
Powering breakthroughs for over 1,200 biotechnology companies, from startups to Fortune 500s
