How to Synthesize your gRNAs for CRISPR
This tutorial is part 3 of 4 in the series “How to Create Knockouts Using CRISPR.” Use Benchling's free molecular biology tools to plan your own CRISPR experiment and design your own gRNAs here.
To edit a cell’s genome, you need to co-express both guide RNAs (gRNA) and the RNA-directed DNA cleaving enzyme Cas9 into the cell. In previous posts, we discussed how to select the best Cas9 for your application and how to optimize the design of gRNAs.
In part 3 of this tutorial, we talk about how to synthesize the gRNAs that we will express in cells to create knockouts. In the next post, we will discuss how to deliver gRNA and Cas9 to cells.
Introduction
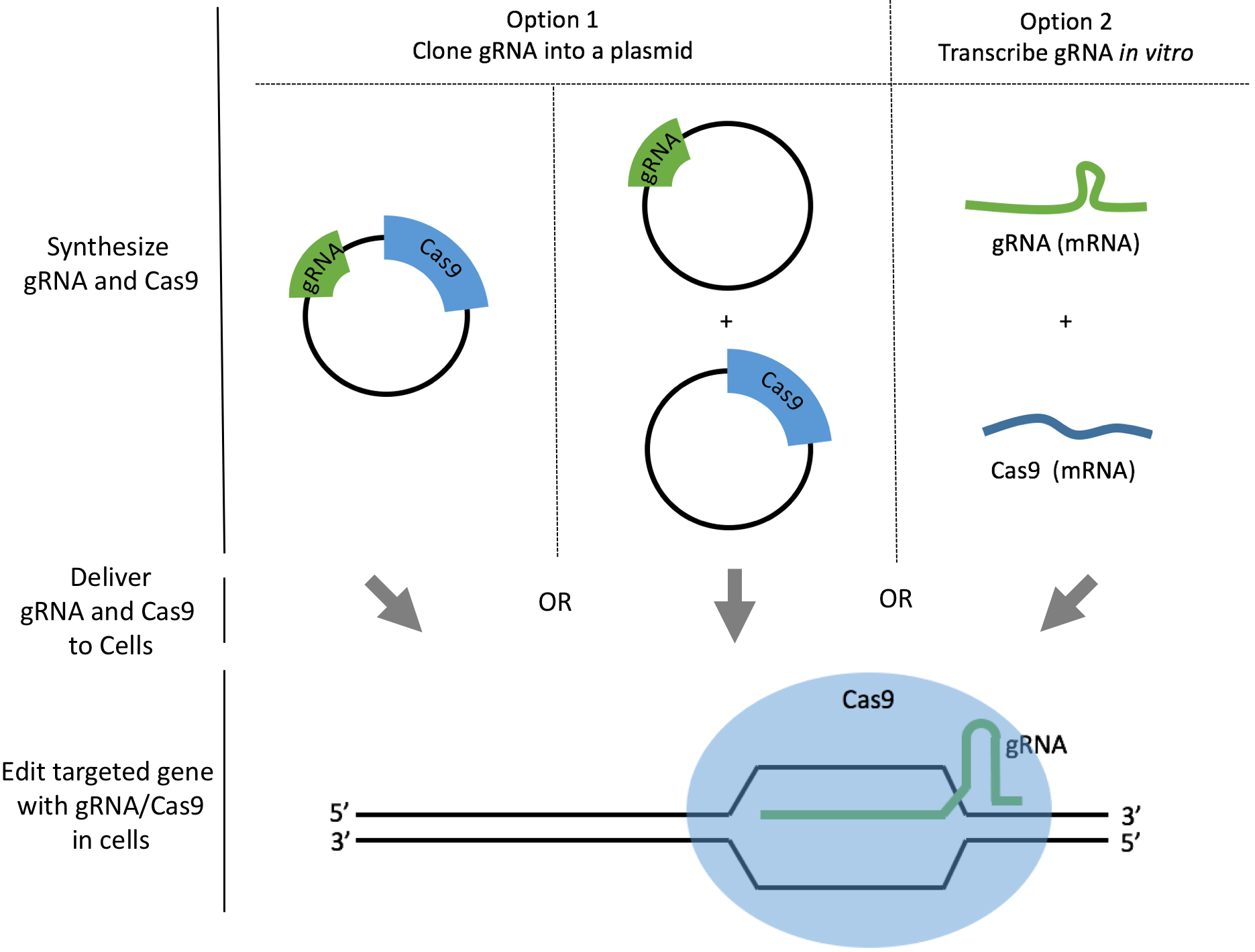
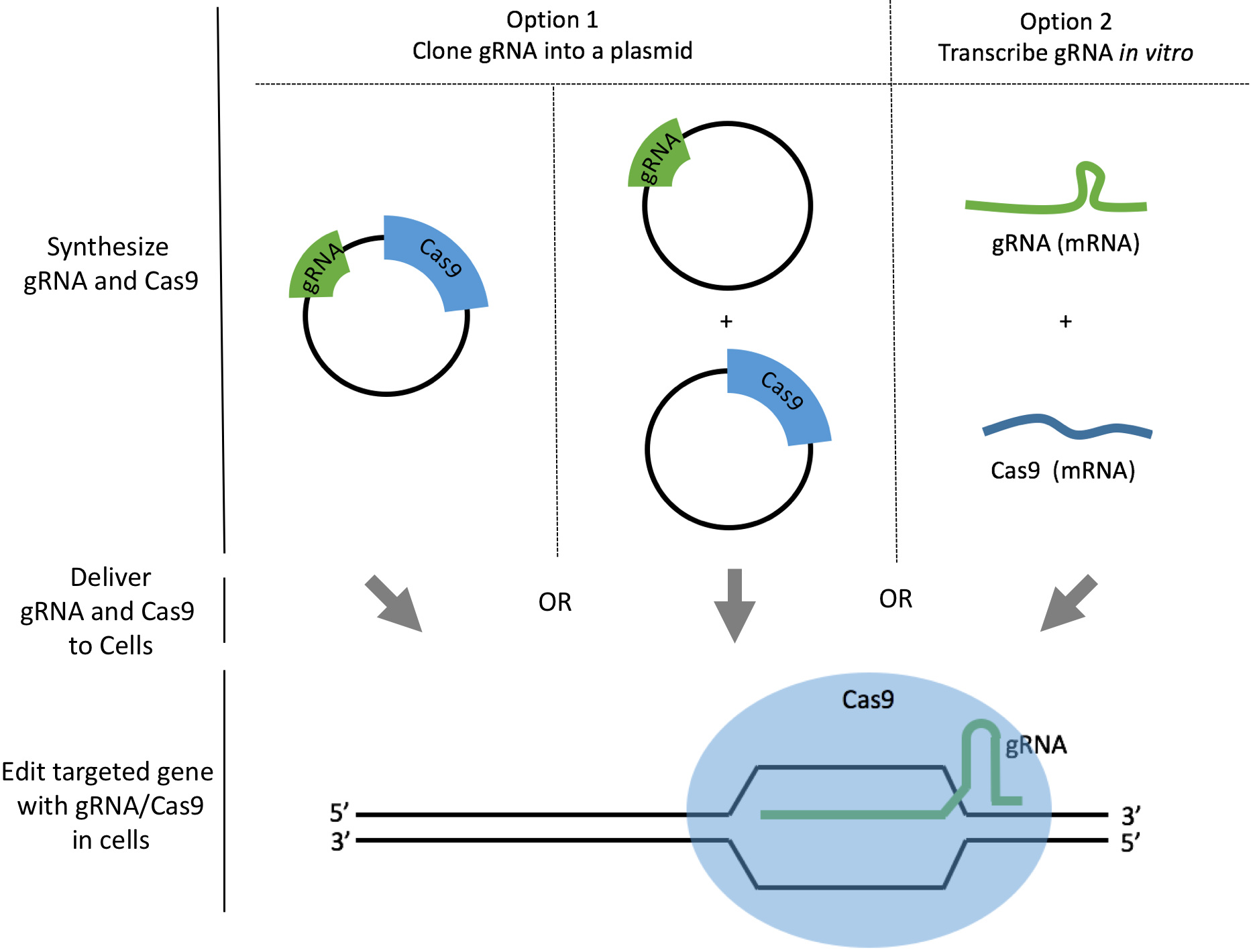
There are two common ways to synthesize your gRNAs: as plasmids or as mRNAs. We will discuss how to synthesize gRNAs as plasmids or as mRNAs and analyze the pros and cons to help you make your decision. For both options, we also link to a comprehensive step-by-step protocol so you can download or edit directly within Benchling. The best method depends on the cell line you work with, and the degree of delivery efficiency required.
How do you know which method works the best for your cell line? It can be hard to search for information related to a particular cell line.
Option 1. Clone gRNAs into plasmids
Researchers have streamlined the workflow for cloning gRNAs into plasmids. Now you can digest the plasmid and ligate the insert in just a single step. We will walk through how to clone a gRNA into a plasmid (PX330) using a protocol developed by Dr. Feng Zhang’s lab at MIT.
Pros
Easy-to-transfect cells, such as human embryonic kidney (HEK) cells, take up supercoiled DNA well, so delivering gRNAs as plasmids usually lead to high transfection efficiency. If you work with easy-to-transfect cell lines, this method can work well for you.
The plasmids are easy to amplify and suitable for long-term storage.
Cons
It is hard to deliver gRNA as plasmids into some cell lines, such as human embryonic stem cells. Researchers have shown that the delivery efficiency is higher in some cases if the gRNA is delivered as mRNA rather than as a plasmid.
Tip
Benchling has a tool to design the primers automatically if you want to clone the gRNAs into a plasmid with BsaI, FokI or other Type IIS restriction enzymes. Check out the tutorial here.
Some plasmids, such as PX330 and lentiCRISPRv2, also encode Cas9, so you only need to deliver a single plasmid to co-express gRNA and Cas9 in cells.
If you will use the same gRNA for other purposes, such as activating or interfering endogenous gene functions, you might want to use one plasmid to express gRNA and the other plasmid to express Cas9.
Protocol
Step 1: Order oligos to synthesize your gRNA.
Step 2: Phosphorylate and anneal each pair of the oligos (1 hr).
Step 3: Linearize the desired vector with BbsI (BpiI) and ligate oligos (2 hr).
Step 4: Transform the final product (1 day).
Step 5: Pick colonies and validate the sequence of each colony (1 day).
What’s next?
You can deliver PX330 by lipofection or electroporation. Alternatively, you can use lentiCRISPRv2 as the vector for viral transduction.
Option 2. Transcribe gRNAs in vitro
In vitro transcription is fast and straightforward once you learn the tricks to working with RNA. The strategy is to add a T7 polymerase promoter upstream of the DNA template of your gRNA. You can then use T7 polymerase to synthesize your gRNA in the form of mRNA.
Pros
If your cells are hard to transfect or do not divide, the delivery efficiency is often higher when gRNA is in the form of mRNA compared to when it is in the form of plasmids.
RNAs are short-lived. Therefore, if you deliver gRNA and Cas9 in the form of mRNA, they will be degraded quickly and will not be inherited by daughter cells.
Cons
If you have never worked with RNA, you might need a little practice to obtain high-quality RNA.
There are no selection methods you can use to screen for cells with gRNA and Cas9.
Protocol contributed from Stephen Floor at Jennifer Doudna's lab
Step 1: Order oligos to synthesize your gRNA.
Step 2: Synthesize gRNA as mRNA in vitro(6 hr).
Step 3: Purify and validate the quality of mRNA (2 hr).
What's next?
mRNA can be delivered to cells by lipofection or electroporation. If you decide to deliver gRNA as mRNA, you might also want to consider delivering Cas9 in the form of mRNA or protein. Delivering Cas9 as mRNA or protein has increased the efficiency of generating knockouts in some cells2
Summary
Now you know the pros and cons of synthesizing a gRNA as a plasmid (option 1) or an mRNA (option 2). Here we summarized what forms of gRNA and delivery methods researchers have tried for some of the most common cell lines. The indel frequency is a proxy for the delivery efficiency of gRNA and Cas9:
Coming up
After you synthesize your gRNAs, it’s time to deliver gRNA and Cas9 to your cells!
Want to use Benchling's CRISPR tools at your company?
References
Yang, Hui et al."One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering." Cell 154.6 (2013): 1370-1379.
Liang, Xiquan et al."Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection." Journal of biotechnology 208 (2015): 44-53.
Shalem, Ophir et al."Genome-scale CRISPR-Cas9 knockout screening in human cells." Science 343.6166 (2014): 84-87.
Cong, Le et al."Multiplex genome engineering using CRISPR/Cas systems." Science 339.6121 (2013): 819-823.
Koike-Yusa, Hiroko et al."Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library." Nature biotechnology 32.3 (2014): 267-273.
Powering breakthroughs for over 1,200 biotechnology companies, from startups to Fortune 500s