Illuminating cell biology: a how-to guide to optogenetics
This post was contributed by Michael J. Harris, a PhD student in Immunology at the University of Cambridge. Michael is in the process of developing an optogenetic tool kit for the interrogation and modulation of signalling events downstream of the TcR (with future applications in different signalling pathways and cell types).
Introduction
The light is shining! A revolution in cell biology is beginning that is changing the way we control protein expression and activity. Although we have known about photoresponsive proteins since the 1970s, only recently have researchers begun to co-opt their use for experimental purposes [1]. Termed optogenetics, biologists use light to control various molecular events with high temporal and spatial resolution.
The first protein in the optogenetic toolkit was the UV-responsive, algal-derived Channelrhodopsin-2 (Figure 1). In 2005, researchers used Channelrhodopsin-2 to activate neurons in vitro. Remarkably, only two years later this protein was used in freely moving mammals [2], an impressive technical feat considering the mice had to be tethered to a light source via an intracranial fiber-optic cable.
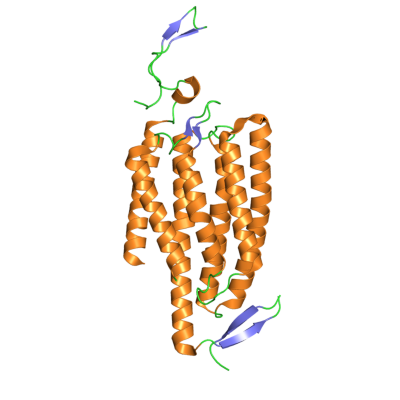
Figure 1. Crystal structure of the Channelrhodopsin-2 photoreceptor
Since these initial studies in neuroscience, we now recognize how optogenetics can impact the cell signaling field. Light-activatable systems lend themselves to cell biology for several reasons:
Unlike pharmacological systems, optogenetic systems can be genetically encoded and stably transduced into your target cell, typically leading to fewer off-target effects.
Unlike traditional knockout or knock-in systems, perturbation to the wildtype system is only induced on light modulation. Thus, cells have less opportunity to upregulate compensatory pathways.
Optogenetics provides spatial and temporal control over signaling pathways in currently unseen ways [3]. It is increasingly clear that cell signals are context dependent, with many cellular receptors sharing the same canonical intracellular pathways (e.g. MAPK and JAK/STAT pathways) [4,5]. Optogenetics provides a mechanism to delineate the distinct aspects of these pathways.
In this post, we hope to give you a taste of the optogenetic applications in cell biology as well as outline a few basic considerations for designing your own optogenetic system.
Selecting Your System
Optogenetic systems can be broadly characterized into two main groups based on their mechanism of action [3]. Before beginning your research, it is advisable to review the characteristics of your system and decide what type of mechanism will work best for you.
Mechanism 1: Light inducible oligomerization
Light inducible oligomerization is also called light inducible separation [6]. In this mechanism, light induces changes in protein structure that alter its ability to bind other proteins or structures. Strickland et al. describe an example of this system, termed TULIPs (TUnable, Light-controlled Interacting Protein tags). which is based on the LOV2 (Light-Oxygen-Voltage) domain (Figure 2) [7]. In this system the Jα-helix of a LOV2 domain from the common oat Avena sativa was engineered with a peptide sequence that undocks from the core domain on exposure to blue light. This exposes a binding site for an engineered PDZ domain. Petra van Bergeijk et al. cleverly applied this system to ask how organelle position affects cellular function by tethering the LOV domain and the PDZ domain to various cellular organelles and cellular motor proteins such as kinesin, dynein, and myosin [8]. No other method to date allows for this kind of non-invasive, fine-scale control of organelle positioning [9].
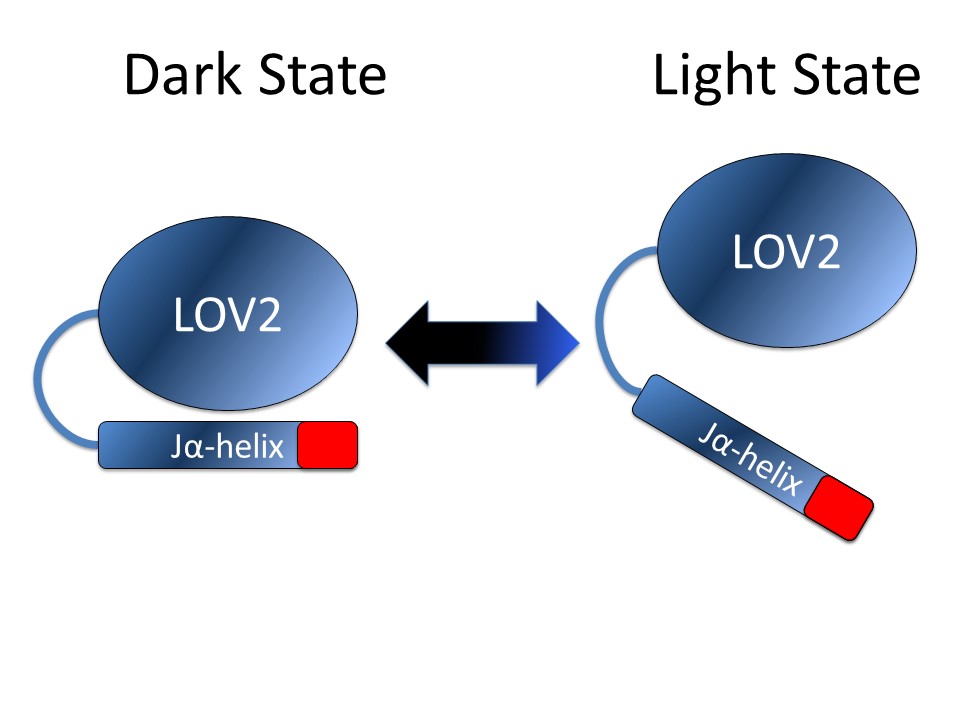
Figure 2: Upon light stimulation the Jα-helix of the LOV2 domain unwinds and undocks from the core of the structure exposing the target sequence (red). Once the stimulus has been withdrawn the LOV2 domain thermally reverts to the dark state within several seconds, allowing for non-invasive, fine-scale control of organelle positioning [9].
Mechanism 2: Photocaging
In photocaging, light can be used to trap (or release) specific molecules or domains. Again using the LOV2 domain as an example, Niopek at al. engineered the Jα-helix of the domain with nuclear localization sequences (NLS) of varying strengths. When fused to signaling molecules such as CDK1 or Cyclin B1, upon stimulation with blue light, the helix is undocked and the NLS become exposed (Figure 2, 3), resulting in nuclear translocation of CDK1 or cyclin B [10]. This system was used to look at the dynamics of cell cycle control with high temporal and spatial resolution.
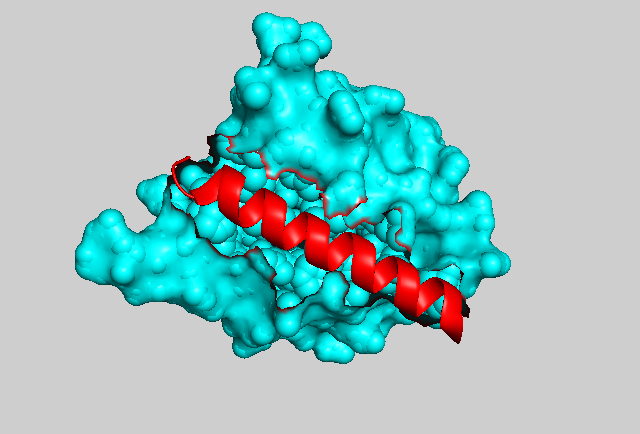
Figure 3: The crystal structure of the Avena sativa LOV2 domain. The core globular domain is shown in cyan and the Jα-helix is highlighted in red.
A summary of many of the systems available can be found in Table 1.
Table 1: The optogenetic cell signalling toolkit.
*This table is abridged and modified from Zhang and Cui’s 2015 review on optogenetic tools [3] with considerations for the recently developed second generation systems. The review itself provides an excellent starting point for setting up an optogenetic system and addresses some of the caveats that come with each of the systems outlined above.
For further reading as you delve into this world of optogenetics, check out these resources:
Pathak et al. [6] have directly compared several of these systems, summarizing the strengths and weaknesses of each optogenetic tool. In their review, they focus on the optical oligomerization systems: CRY2, TULIPs and PhyB. They highlight the size of the proteins, their effectiveness at activating simple genetic switches, and their background activity.
Taslimi et al. characterized second generation CRY2 constructs [11]. In their paper they apply the CRY2 protein in a photoactivatable Cre-recombinase reporter system. The paper addresses some of the issues including background activation and clustering that were observed in previous studies.
The overall power of optogenetics is excellently exemplified in Toettcher, Weiner, and Lim’s cooption of the PhyB system to interrogate Ras/Erk signaling [12]. Here, they apply different patterns of light stimulation to vary signal output responses using a photo-activatable MAP kinase. Their research bridges the fields of engineering and biology to look at the cell as a ‘black box system’.
Zhou et al. describe the premise of the Dronpa protein system by using cyan light to modulate protein degradation in mammalian cells [13].
The Devil is in the Details
Once you have decided on a mechanism and a light-responsive domain, you can begin to work out the details of your system. There are still several important factors to consider:
Fluorophore tags:As most of the optogenetic domains come from naturally occurring proteins, they tend to respond to quite a wide spectra of light [14,15]. This may complicate any fluorescent microscopy you plan on performing. For example, the LOV2 domain responds to 450nm blue light, but overlaps with the excitation spectra of both BFP and GFP (405nm and 488nm, respectively). Thus, if you use the lasers to look at GFP or BFP, you will inadvertently activate the LOV2 domain. (The same can go for focusing the microscope using white light!) Keep in mind that it is considerably easier to swap out fluorescent tags than it is to set up a new optogenetic tool.
Chromophores or co-factors:Many of the light-inducible domains rely on a chromophore or co-factor for function [14]. Some co-factors, such as FMN (Flavin mononucleotide) and FAD (Flavin Adenine Dinucleotide), are expressed in many cell types and/or tissue culture medias [16]. Others, such as PCB in the PhyB system, are not, and therefore require supplementations.
Binding kinetics and rates:You can tweak the binding kinetics and rates of your optogenetic system through several means. The addition of a drug such as imidazole, the characteristics of the binding partners as described by Strickland et al. with the TULIPs system [7], or point mutations can alter the rate at which optogenetic domains photocycle [17-19].
Controls:As a control, it is best to include a photo-nonresponsive mutant version of your protein. The UniProt websiteis an excellent resource to help identify these mutations. Many of the other mutations have also been well-characterized elsewhere [6,20-21].
Conclusion
The use of optogenetics in molecular and cellular biology is only just beginning. As the movement takes hold, we will begin to see more creative and insightful uses for these light-controllable tools. Although there are no plug and play systems currently available, don’t be intimidated about setting up your optogenetic experiment! Go forth and see the light!