How to Design Homologous Recombination Template for CRISPR
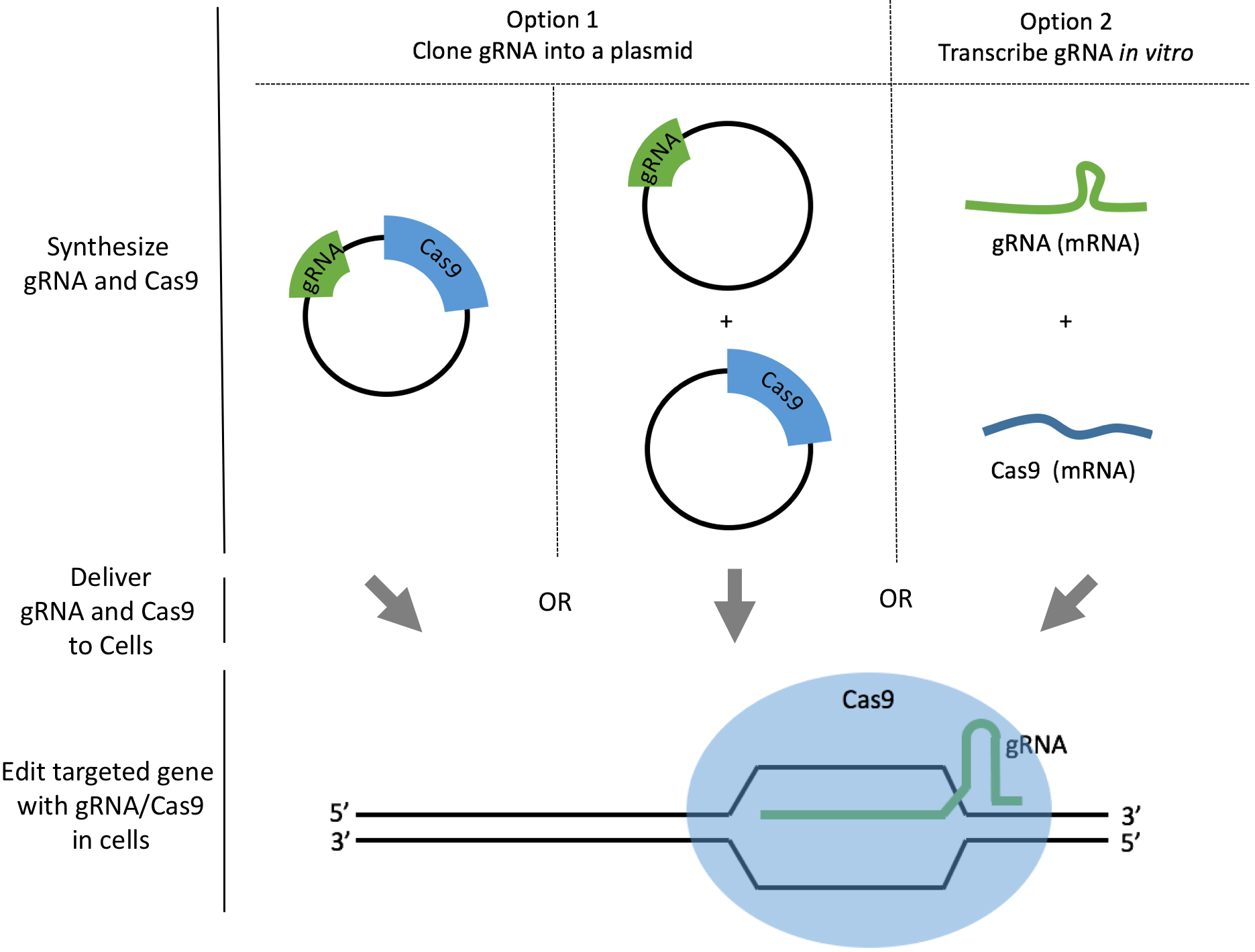
Using CRISPR to perform precise genome editing is efficient and relatively simple [1], [2]. You first use gRNA coupled with Cas9 to create a DNA double-stranded break near the genomic region you want to edit. Then, you design a DNA template to introduce your desired mutations into the cell’s genome. The cell can repair the double-stranded break with homology directed repair (HDR) via homologous recombination (HR) mechanism. Therefore, the DNA template is also called HR template or HDR template.
So how do you design a good HR donor template? Designing the template manually could be time-consuming. Luckily, Benchling has built a wizard to help you design your HR template as single-stranded oligodeoxynucleotides (ssODNs) for introducing small changes (< 50 bp) in three steps. Here we will show you how to use the wizard and discuss important factors to consider during the process.
Before you start
First, you want to design gRNA so the cut site will be within 10~30 bp from the targeted genomic region [3][4]. Check out this post to learn some tips for designing gRNAs with high specificity.
Step 1: Create the HR template and design modifications
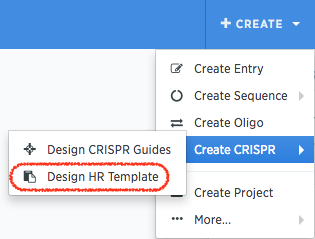
First, in Benchling, start by clicking “Create” in the top right. Under “Create CRISPR” you can click “Design HR template” to open the HR template wizard.
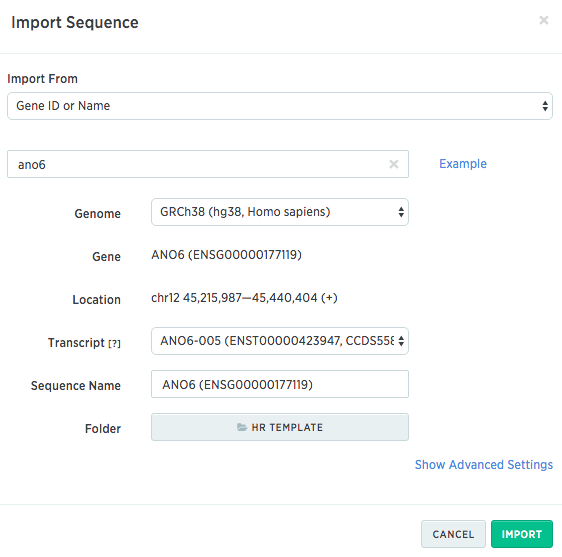
In the next dialog box, you can search for your gene of interest. Here is what it looks like to search for Ano6 in a human genome (hg38):
Then you will need to provide the information of what genome and PAM sequence you want to use to design HR template:
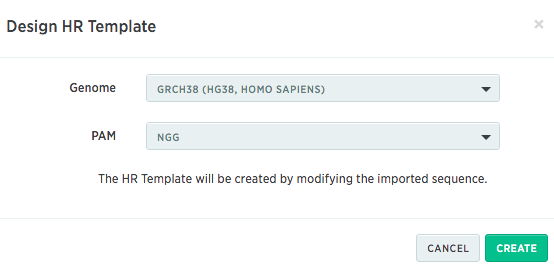
After you import the sequence map, the HR template wizard will ask you for the mutations you want to incorporate in the template. All the edits you make will be recorded on the right panel.
For example, here I made a point mutation from T to A, so the new 60th amino acid will be Glutamine (Q) instead of Leucine (L).

Tips
If you want to design the HR template from an existing sequence on Benchling, you can click the crosshair icon on the sidebar to start the HR template wizard.
For the most effective HR template for CRISPR, make sure the mutations you introduce are within 10~30bp of the cut site.
This wizard is designed for introducing small changes (100 bp), be sure to manually check if there are additional Cas9 target sites in your template beyond the single site found in Step 3.
Step 2: Adjust the length of HR arms on both sides of the mutation
In the next step of the HR wizard, you can adjust the length of HR arms on both sides by highlighting regions of interest on the Sequence Map. In this example, I made 75 bp HR arms on both sides of the mutation, resulting in a 151 bp ssODN HR template:
Tips
The size of your intended mutation will determine (1) the ideal length of HR arms and (2) whether you should use ssODNs or plasmid donors as the basis of your HR template.
For small changes (<50 bp), it is most efficient to use an ssODN as the HR template. In this case, create 50-80 bp homology arms that flank each side of the changes [5].
If you want to introduce larger changes (>100 bp), it is more efficient to use a plasmid donor than an ssODN. In this case, it is best to design homology arms that are around 800 bp that flank each side of the the changes. After designing the HR template, you will need to clone the template into another donor vector [6].
Step 3: Incorporate mutations in the target site of the HR template (optional)
A common glitch in doing Cas9-mediated gene editing is when the HR template contains an intact Cas9 target site/PAM site. In this case, Cas9 may bind to the target site and degrade the HR template. Additionally, even after successfully introducing the mutation with the HR template, Cas9 may continue to cut the genomic site if the Cas9 target site and PAM site were left intact. To avoid the situation, you can use the next step of the HR template wizard to mutate the target site to avoid any unintended consequences.
To start, copy your guide sequence in the HR template wizard:
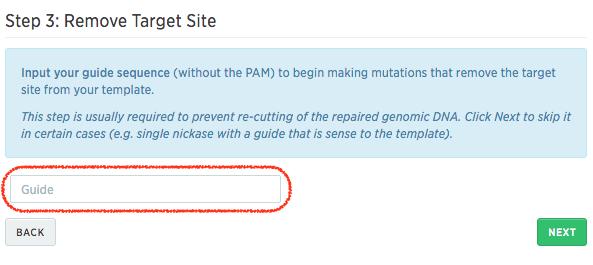
Once you input your guide, Benchling will show you a table of possible silent mutations to change the target site in your HR template:
To avoid the HR template being degraded by Cas9, it is most effective to mutate the PAM sequence. The wizard will automatically select a good mutation for you.
In this example, since the target sequence is within a coding region of the gene Ano6, I want to make sure the mutation is silent and the codon adaptation index (CAI) similar to the original sequence. The table shows me that I can introduce a silent mutation at the PAM sequence if I change CCG to CCA, CCT, or CCC. The codon adaptation index of CCA is closest to the original sequence. However, the alteration would make the PAM sequence to be CAG, which still has affinity to for Cas9. Therefore, the wizard suggests that I should mutate CCG to CCT instead.
Now that you are done with all three steps of the design wizard, you are ready to synthesize the HR template!
Tips
To achieve high knock-in efficiency, it is best to synthesize a high-quality ssODN, such as ultramer oligo or non-PAGE purified oligo from IDT.
Some scientists test both the sense and anti-sense HR template to optimize the knock-in efficiency. You can easily copy the sequence and its reverse complement on Benchling.
Conclusion
Counting the length of HR arms and figuring out how to introduce silent mutations manually is labor-intensive. Hopefully, Benchling’s HR template wizard can speed up your design process.
Want to learn more tips for optimizing CRISPR knock-in experiments? Check out this post written by Ralitsa Madsen from the University of Cambridge. She explains how she optimized her CRISPR knock-in strategy to get an efficiency of 18% in HEK293s cells with a wild type SpCas9.
Curious about different strategies for synthesizing gRNA? Check out this post with an in vitro transcription protocol from Stephen Floor in Jennifer Doudna’s lab!
References
Cong, Le et al. "Multiplex genome engineering using CRISPR/Cas systems." Science 339.6121 (2013): 819-823.
Mali, Prashant et al. "RNA-guided human genome engineering via Cas9." Science 339.6121 (2013): 823-826.
Yang, Luhan et al. "Optimization of scarless human stem cell genome editing." Nucleic acids research 41.19 (2013): 9049-9061.
Singh, Priti, John C Schimenti, and Ewelina Bolcun-Filas. "A mouse geneticist’s practical guide to CRISPR applications." Genetics 199.1 (2015): 1-15.
https://www.addgene.org/crispr/zhang/faq/
Yang, Hui et al. "One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering." Cell 154.6 (2013): 1370-1379.
Powering breakthroughs for over 1,200 biotechnology companies, from startups to Fortune 500s